SEGYJp2 Version 1.0 (DRAFT VERSION)
Bob Courtney
Geological Survey of
Natural Resources
Bob.courtney@nrcan.gc.ca
March 30, 2009
Table of Contents
SEGYJp2
Version 1.0 (DRAFT VERSION)
Intellectual
and Property Rights
Main
Tab: Adding files to the Input Process List
Spectra
Tab Page : Exploring Input SEGY File
Convert
SEGY to SGYJP2 Tab Page:
Appendix
I - Storage and XML Schema
Routines to Manipulate JPEG2000 Box Data
Introduction
The image is the data and the data is the image.
SEGYJp2 is a Windows XP program that implements a codec to convert SEGY data to and from an extended wavelet-based JPEG 2000 storage framework.
Although. JPEG 2000 is commonly used for images is possible as well to store variable bit length bipolar data.
SEGY-encoded JPEG 2000 files will viewed simply as a grayscale image in most commonly available JPEG 2000 viewers (e.g., Adobe Photoshop, LizardTech, Corel draw, etc.) . (Note resample SEGY data in the conversion process to 8 or 16 bits for compliance to some viewers)
However, these files contain much more, they contain all the trace information derived from the SEGY header data encoded as XML,and stored directly in the file. This information can be recovered and reconverted back into the original SEGY. An enhanced viewer that uses these extended capabilities is available through the GSC.
It is important to recognize that the trace data from the SEGY file are stored directly in the JPEG 2000 as either unipolar or bipolar data and not, as many would suspect, as grayscale image-type representations. Although these data are viewable as image, the actual trace amplitudes are stored natively in the file as integer values and can be extracted from the encoded JPEG 2000 file.
This program represents the encoding component of a linked processing flow for large volume, high resolution SEGY data implemented at the Geological Survey of Canada.
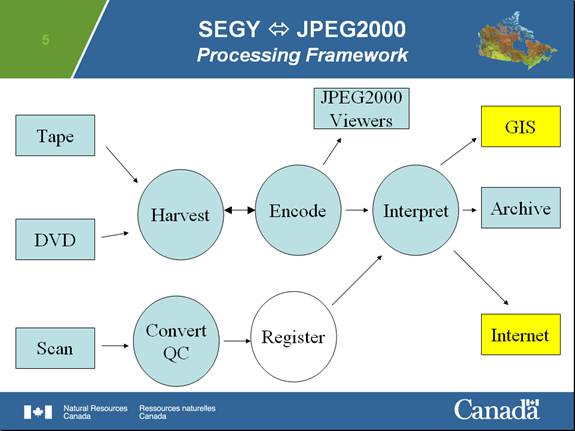
The attempt here is to implement a database- and internet-ready framework that will allow researchers and others to easily exchange and archive both sonar data and their interpretations. Our experience has suggested, that at least for single channel high-resolution seismic and sidescan data, scientists much prefer working with these JPEG2000-based digital sections rather than with the original SEGY data in industry-standard seismic mapping and display programs.
JPEG2000 encoding of seismic and sidescan offers the capability to encode SEGY data efficiently (typical compression of 10:1 though to 40:1), offer quick evaluation of data quality through off-the-shelf image viewing technology and, importantly, afford a framework for embedding value-added interpretations to accompany the data. These files may contain 100,000’s of traces efficiently without undue allocation of system resources.
The adoption of a standard, open structure framework for storage allows this work to be leveraged by other technologies developed for dissemination of imagery stored in JPEG 2000, such as image servers and similar types of technologies. For example, the development of the JPIP internet transport mechanism for efficient transfer of JPEG 2000 data over serial communication lines holds great promise for interactive viewing and manipulation of these large data objects over standard TCP/IP connections.
One of the notable advantages of this approach is the multiresolution or multiscale nature of the JPEG 2000 storage framework. In storing the data in a multi-resolution form, near-optimal images of these data can be displayed as one zooms in and out with very little effort and very little load on computer resources. This feature makes the onscreen representation of the seismic data very similar to what one would see viewing a printed seismic section, with the added advantage of course that these data and the related interpretations are geographically registered.
JPEG2000 uses an efficient entropy –based MQ bitstream encoder for data storage. Therefore white space compresses very efficiently (as it has no entropy). This feature allows us to store seismic data zero-padded for trace delay changes, yielding seamless imagery which would be prohibitive in size in an uncompressed form. On the conversion of JPEG2000 files back to SEGY, these zero-padded section are removed, recovering the original trace delayed form.
Encoding implemented
in SEGYJP2
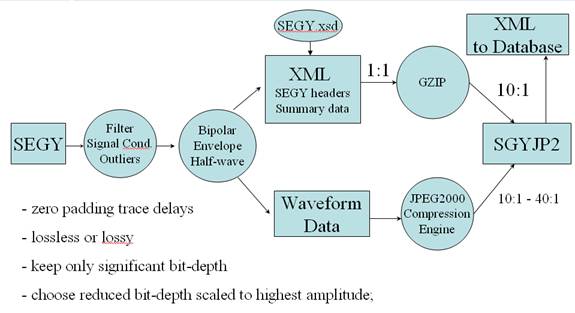
This program implements the processing flow sketched out in the figure above:
In the first step, a SEGY file is optionally bandpass-filtered, signal conditioned, and outliers removed.
Then the choices made to save the data in the original bipolar form or in a mono-polar form (envelope detected or half wave rectified).
The trace data in the SEGY file is compressed with the JPEG 2000 compression engine at a specified bit depth ( ranging from 1 to 32 bits, the choice depending on application or the original resolution of the data). The data are rescaled to the chosen bit depth such that the maximum amplitude in the conditioned file has full bit depth span. In this way, the entire bit depth is fully utilized.
Intellectual and Property Rights
This
program is the intellectual property of the Government of Canada. All rights to
modify and distribute this software are retained by the Government of Canada.
"Intellectual
Property" means all rights in respect of Confidential Information, any
know‑how, inventions and improvements related to Confidential
Information, including without limitation, patents, copyrights, trade secrets,
trade‑marks, registered industrial designs, any applications for same and
all rights therein.
"Confidential
Information" means any information of a scientific or technical nature
disclosed to, generated or acquired by the Student in the course of carrying
out the Student activities defined in this Agreement, whether oral or recorded
in any form or medium and whether or not protectable by copyright.
We allow this program to be redistributed
freely in its original form. However, please send me an email if you have a
copy – I’ll ensure you get on a mailing list for upgrades.
Contact
the author, Bob Courtney, Natural
Resources Canada,
Unfortunately
we have limited capacity to support needs of outside users. We do however encourage
joint research agreements to extend this framework.
The SEGYJp2 distribution package may be freely distributed as it contains no proprietary code. It does utilize Kakadu (www.kakadusoftware.com) , GSL (http://www.network-theory.com/gsl/) and TChart ( www.steema.com) libraries which are distributed in compliance with each of their respective licensing agreements.
The product will undoubtedly evolve over time so it is prudent to
contact the author to obtain the most recent version. Since this product is
free, sometimes one must wait until time is available to address bugs, etc.
This software was developed as part of the Geoscience for Ocean
Management Program as a means of viewing and disseminating high-resolution
digital seismic and sidescan data collected though marine operations of the
Geological Survey of Canada.
Installation
SEGYJp2is easy to install.
(a) If you are within the Natural Resources Canada network, you can install it directly from a network drive. Open a window to \\s5-dar-data1\shared_software\SEGYJp2\setup.exe .
If you are outside GSCA but within the NRCan network , try using this link: \\192.55.224.44\shared_software\SEGYJp2\setup.exe
Proceed as is typical for any Windows application installation. A Canadian Flag icon will be added to your desktop and an entry under the folder NRCan will be added to your Program list.
(b) If you obtain a ZIP file containing the release, unpack the zip file to a directory on a local drive and use Explore to enter the SEGYJP2 subdirectory that is unpacked into the specified spot. Double click on setup.exe and proceed as above.
Removal
To remove the package, choose Control Panel from the Windows Start Menu. Double click on Add or Remove Programs. Scroll down to SEGYJp2 and click the Remove button. Follow the prompts.
Note : It is necessary to manually
remove an existing package before a newer version can be installed.
Operation
Startup
To start the program, double click on the desktop SEGYJp2 icon or the icon within the Program Files/NRCan menu entry.
If all goes well, the main window will appear of the desktop ( Fig 1).
Figure 1 - Main Window of SEGYJp2
Convert SEGY to JP2 :
Main Tab: Adding files to the Input Process List
From the menu bar at the top of the page, choose File => Open SEGY Files to add SEGY files to the process list. By default, the file selection box will look for files with a “.sgy“ extension , but that feature can be overridden to include all files in the file selector box.
The program will conduct some rudimentary tests on the
chosen files to ensure that they are SEGY files. Most of the time these tests
detect SEGY compliance- in the odd instance a noncompliant file
can pass through these tests and may well cause
failure of the program.
It is assumed that the source SEGY file contains only one channel of information. That being said, the program will convert SEGY files containing multiple channels with the provision that the trace length and sampling times are the same for every channel. Since this program does not change any values in the SEGY trace headers that are stored in the jp2 file, images of these multichannel files may look somewhat confusing when viewed, a conversion of this multichannel form back into SEGY will recover the original state of the source SEGY file.
The file list may be cleared, or selection within list removed, by using the buttons under the list.
Scanning files
Summary data of the SEGY files added to the input process list can be generated by double-clicking on the file name in the list. :
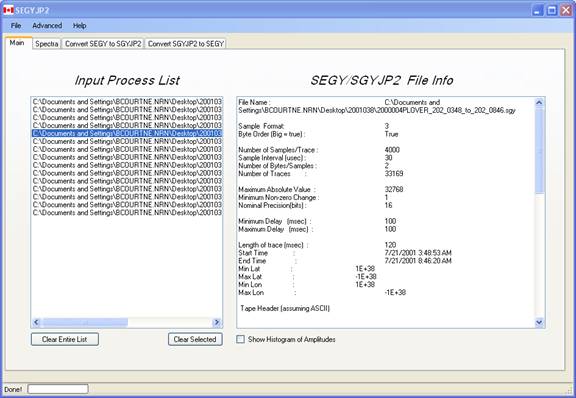
Care must be taken if
variables have been set in the Spectra or Convert SEGY to SGYJP2 tabbed pages
(e.g., band pass filtering, envelope detection). These options are executed
when the file is read such that the summary data will reflect these chosen
processes.
If the show histogram of amplitudes checkbox is enabled, a plot of the CDF of the data against normalized maximum amplitude is generated. This plot is useful for determining outlier properties.
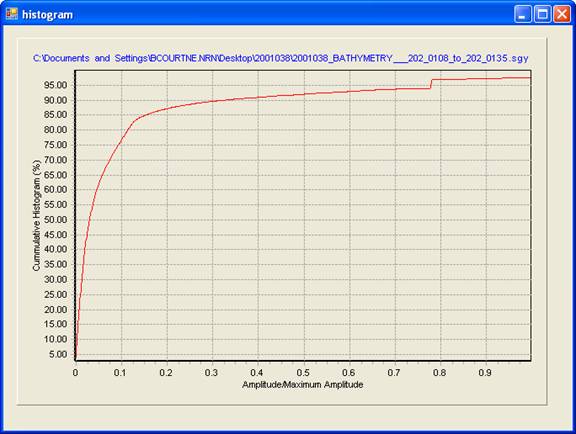
This plot can be panned by holding the right mouse button down and dragging the image. This plot can be zoomed by holding the left mouse button down and dragging a rectangle from its upper left corner to a lower right corner. The view may be reset by dragging a rectangle in the reverse direction, from the lower right corner to the upper left corner.
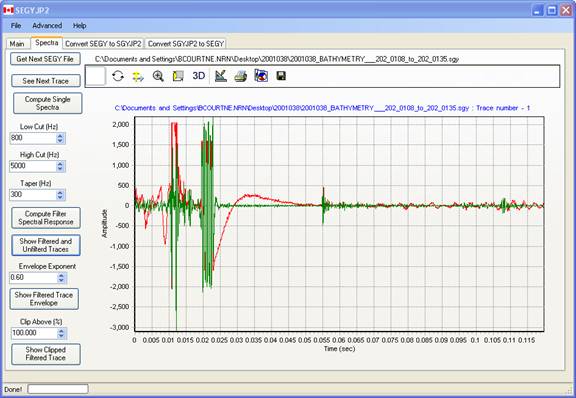
Spectra Tab Page : Exploring The Input SEGY File
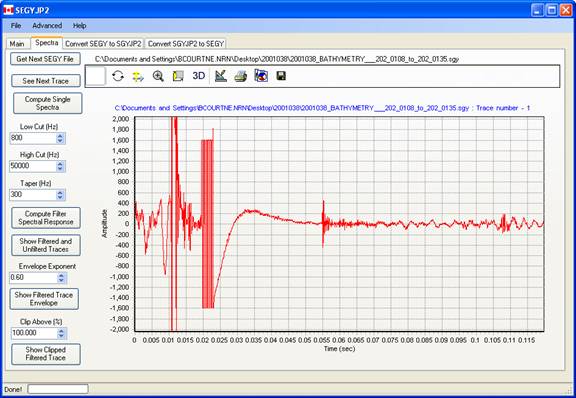
The Spectra tabbed page allows the user to visualize the trace data contained within the SEGY file and also to assess the effect of processing parameters. By default, the data in the selected file in the input process list is displayed, or the first file in the list if no file have been chosen.
The toolbar located above the graph is a standard plug-in from the graphics module, TChart (www.steema.com). This allows the most features of the plot to be customized, data within the plot saved as text, XML, HTML, or Excel format, and the plot to be saved in a variety of graphic formats, copied to clipboard, printed, etc. . It can even used to rotate the plot in3-D ( why one would do this, I don't know! ). Please play with the settings.
![]()
Get Next SEGY File : choose the next file in the input list.
See Next Trace: move to the next trace in the current SEGY file
Compute Single Spectra :
This button calculates and displays a single FFT spectra of the current trace. No averaging of successive trace spectra is performed, nor is any spectral averaging within the single trace estimate performed. And we all know what this means about the expected variance in the spectral estimates.
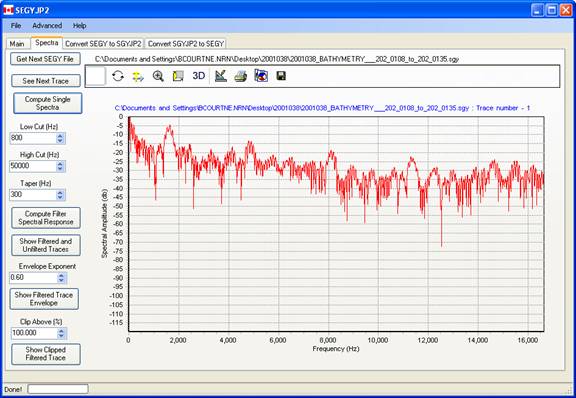
Bandpass Filter Settings :
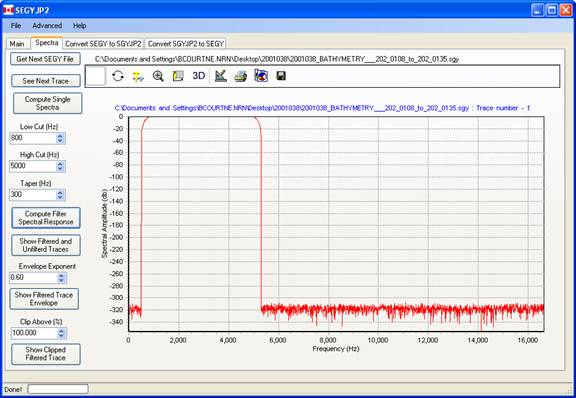
Hi cut and low-cut filter settings are next applied along with a taper length, which specifies the frequency interval over which the filter amplitude varies from 0 to 1. The filter is implemented as a zero phase, mixed radix FFT-based filter. Future plans include different filter tapers for the low and top ends.
If the hi cut value is greater than the Nyquist frequency for the times series, then the hi cut value defaults to the Nyquist.
Compute filter
spectral response:
The spectral response of the band pass filter may be displayed by pressing this button.
Show Filtered and
Unfiltered Traces:
This button is used to plot the input SEGY trace (red) along with the trace filtered by the specified bandpass (green). This allows the user to graphically see and appreciate what the bandpass filter is doing to the data.
The user can zoom in two phases of interest by rubber-banding a rectangle on the plot from top left to bottom right, holding the left mouse button down . The plot extents may be reset by rubber-banding in the opposite direction
Please refrain from
using the magnifying glass in the toolbar, this option is used to change 3-D
viewing perspective. If by chance you happen to put the plot in 3-D mode,
toggle the 3-D button on the toolbar to return to 2-D mode.
Envelope detection:
Often with broad band sources ( e.g., high-resolution boomer data), echo sounder type sources ( e.g. . Knudsen chirp) or high noise to signal situations, the display of data is improved by calculating the envelope of the trace.
Envelope detection in this program is executed through a convolution-based approach.
After we calculate envelope of the trace data, we apply a transform of the form
![]()
where x is the envelope detected amplitude, y is the transformed variable, and α is the envelope exponent.
We apply this transform for two reasons:
- The transform applies some amplitude compression such that grayscale visualizations of the data show detail in both high and low amplitude parts of the section.
- More importantly, if the original trace contains random environmental or other kind of noise with the normal distribution, then the envelope of this noise would have a Rayleigh statistic.
A Rayleigh statistic is what statisticians call a heavy-tailed distribution; that is, there are relatively numerous instances of high amplitudes; these effects manifest as speckle in images ( a common characteristic of SAR type imagery).
In the conversion of trace data to wavelet-based JPEG 2000, we prefer a data distribution with a Gaussian statistic as optimal compression and image fidelity is achieved with normally distributed data.
Numerical modeling has shown that a choice of α = 0.6 will transform a Rayleigh distribution to a distribution very similar to that of a non-zero mean Gaussian. Matlab code demonstrating this principle is available from the author.
Envelope Exponent :
Choose a value from 0.4 to 1.0. We recommend leaving the value at the default of 0.6 for the reasons outlined above.
Show filtered trace
envelope:
Click this button to show the transformed envelope of the band-passed waveform data. If the envelope exponent is changed, the display will interactively reflect these changes.
Clipping:
In an ideal world, sonar data is uncorrupted by sound coming from other sources. In practice, however especially in the high-resolution game where multiple sources are often the water, seismic data are overprinted by unwanted high amplitude noise, these noise sources can be both incoherent and coherent in nature.
In JPEG 2000 compression, data can be expressed with variable bit depths ranging from 1 to 32 bits for this implementation. We desire that the prescribed bit depth is used to span the waveform amplitudes of interest; we do not want to precisely define high amplitude spikes or other such data.
We implement histogram-based clipping as a means of removing high amplitude outliers that lay outside the normal variance of our data.
Clip above:
The clip above variable determines the percent of amplitudes in the data set that are passed unaltered into the JPEG 2000 compression engine. Amplitudes above this prescribed value are clipped and set to the maximum value determined by the normalized cumulative distribution function of the data.
In the spectra page, clipping is performed on a trace by trace basis but when we process the whole file, clipping is performed on the global aggregate distribution of data in the input file.
If this clip above value is set to 99%, for example, all data with amplitudes less than the 99% probability of the CDF pass unaltered, but all data values above the 99% CDF level are set to the clip level.
In practice, for envelope data, values of the clip above variable range between 99 and 100%, a starting value of 99.5% is reasonable.
The best way to check this value is to process the file and look at the resultant image. Images of the data, where the clipping level is too high, will appear too washed-out or faint, images of the data or the clipping level is set too low will appear oversaturated.
Show clipped filtered
trace
This button will show the effect of clipping on the currently loaded trace. Changing the value of the clip threshold will interactively display the results on the screen.
Convert SEGY to SGYJP2 Tab Page:

In this stage, the main work of converting all the files in the input list to sgyjp2 is done as a batch process. A typical conversion of a single SEGY file takes about five to 15 minutes so hopefully we can press the start button and come back in a few hours and everything will have been converted.
Expedition Metadata
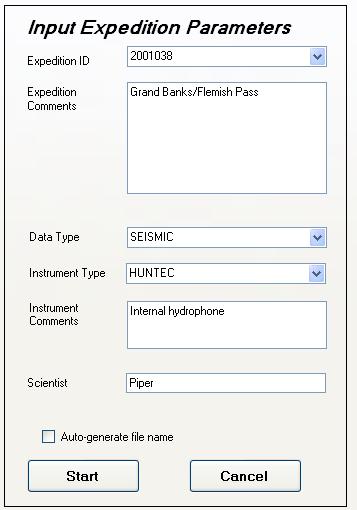
Expedition-specific metadata is entered in the left-hand panel of this page. These data are saved as XML string type variables within the processed JPEG2000 file, and may be later on extracted and used to populate databases to describe compressed file holdings.
At the GSC, we use enumerated variables to describe the expedition ID, the data type, and the instrument type. These enumerated values can be chosen from the drop-down lists associated with each entry box. The expedition ID entry box is not kept up to date so users need to carefully enter expedition ID codes that are not found in the drop-down list.
Although all these enumerated values can be overridden in free-form text entry by the user, they should correspond at best as possible to enumerated or fixed values within our databases. This is a real concern for GSC or NRCAN employees. Proper entry of the expedition ID code for example, is mandatory for subsequent merging of navigation data into the JPEG 2000 files, where the navigation is sourced from our databases.
Compression
parameters:
The compression parameters are specified in the upper right hand corner of the page.
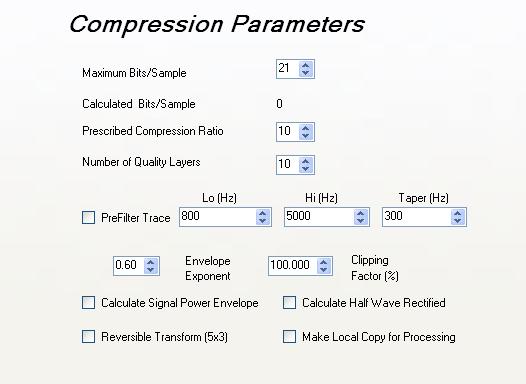
Compression
parameters: maximum bits per sample
JPEG 2000 is not limited to fixed word lengths when storing data. It stores bit planes of the wavelet transform of the data and the number of bit planes can be tailored to best suit the desired resolution in the input data stream.
This numeric entry box specifies the maximum number of bits to been encoded for each trace sample within the input SEGY file. By default, the maximum bits per sample is 21 bits spawning a 126 dB of signal-to-noise - far exceeding any realistic dynamic range of data collected in the field.
SEGY data stored in floating point format have nominally 32 bits of dynamic range. But this is rarely, if ever, utilized within these files. We convert floating point data to integer form to encode these files in JPEG2000. In this case, a floating point number for the maximum amplitude of the input file is found and stored in the JPEG 2000 file, and signal amplitudes are stored in scaled integer format at the prescribed bit rate.
Since this bit range will be applied relative to the maximum signal amplitude in the input file after bandpass filtering and clipping. So that this bit range corresponds to true dynamic range within the significant data.
Storing only 21 of 32 bits of a SEGY file immediately reduces the file size by a factor of 21/32, a 66% compression of the original file without loss above -126 db. If these JPEG 2000 files are subsequently converted back into SEGY format, they will be accurate within the bit depth used in the compression if a lossless compression ratio of 100% had been applied during the compression.
It should be noted that many commercial image viewing packages, such as those offered by Lizardtech and Adobe, may only be able to image data stored in eight bit and 16-bit formats. Choosing the maximum bits per sample equal to eight or 16 will allow the encoded JPEG 2000 files to be viewed as images in these commercial packages. Since 16 bits corresponds to 96 dB of signal to noise and that the 96 dB is applied from the maximum amplitude of the data set, 16 bits affords more resolution than is needed in most cases.
Compression
parameters: calculated bits per sample
The input file is scanned and estimate is made of the bit-depth recorded within the raw data of the source file. If the calculated bit depth in the source file is less than the maximum bits per sample, then the data is stored at the lower bit rate. For example, during the 1990s, our onboard digitization systems recorded 12 bit data and typically this is the bit depth used to store the data.
However if the data is filtered or envelope-detected ( these are floating point operations), the will be converted to floating point data.
Compression
parameters: prescribe compression ratio
The compression ratio is the size of the output JPEG 2000 file divided by the size of the input SEGY file expressed as a percentage. By default, a 10% compression is applied so that the output JPEG 2000 file will be roughly 10% of the size of the input file.
We have found very few artifacts in any data compressed at the 10% level, and we feel that this value is conservative for most seismic data sets. Even the value of 2 to 4% may prove sufficient as it usually produces very little distortion of the seismic wavelets. The most distortion would be noticed at artificial boundaries within the data set, say if the traces were blanked down to a level just above the seafloor.
We would encourage the user to experiment with this setting and find the most appropriate setting for their application
Note: trace delays in SEGY
files are used to generate a zero padded areas of the section so that features that
trend across traces having different delays lineup in the JPEG 2000 image file.
This zero padding effectively increases the raw size of the file and, at present,
the compression ratio is applied to this expanded, padded file size. In these
instances where the trace-delay-padded data file is much larger than the
unpadded SEGY file, one may have to choose smaller values of the compression
ratio to achieve the desired net compression. Hopefully this shortcoming will
be addressed in future versions.
Compression parameters: prefiltering
The bandpass filtering strategy described in the previous section is applied to files on the stage during reading the input SEGY files. The values applied in the spectra page should have been transcribed into the numeric entry box is shown on this page. But to be safe checked to make sure these values in the high-cut, low-cut, and taper input boxes are appropriate. The bandpass filter is applied if the check box labeled prefilter trace is enabled.
Compression
parameters: envelope exponent
The envelope exponent is applied to all band passed and clipped data.
I would try running the program using a value of 0.6 for reasons mentioned in the previous section on spectra. Although this rationale was based primarily on statistical characteristics of the envelope detected data, this exponent has proved useful simply as a signal conditioner that helps display dynamic range within the data set.
Compression parameters:
clipping factor
The clipping factor is applied to remove outliers within the data set as a whole. For envelope detected data. A value of 99.5% or even higher seems to give good images of envelope-detected seismic data in JPEG 2000 viewers. I find lower values of between 95 to 99% are more appropriate for bipolar and half wave rectified versions.
So try running the program using one of these values. And if the resulting image on inspection is too dark, then increase the clipping factor. If the resulting image is too faint in areas of interest, decrease the clipping factor. You'll soon get an idea of how the clipping factor affects image fidelity.
In the SEGYJp2_Viewer program, one can change the palette of the image, but we attempt here to choose an appropriate clipping factor such that, on first view, the data looks well balanced in a tonal sense. Often the first view is what will be seen by those scanning your inventory through network-based image viewers ( e.g., Lizardtech).
Compression
parameters: envelope detection
Check off this box if you want the envelope-detected version of the input SEGY file. We find that the envelope detected waveform is highly appropriate for high-bandwidth boomer type data and also 3.5 kHz chirp type sounder. The resultant file from this choice has an “_envelope” suffix added to the root name of the input file name.
Important to note here
is that whenever we take a bipolar waveform and transform it into a unipolar form, whether is be an envelope- detected or half
wave rectified signal, the fundamental spectral content of the information
changes.
Unipolar data has non-zero mean values whereas bipolar
data tends to average out to zero. JPEG 2000 encoding of the data can be viewed
as a series of nested averaging and differencing operations. As one zooms out
of the JPEG 2000-coded image one looks at the averages of the data. When one zooms in, zero-mean differences are added back into the
average yielding more detail.
Therefore, if one
zooms out on a unipolar-encoded seismic image, they would observe an
averaged picture with progressively less
detail – much as would be observed as an observer backs away from a printed
section.
However, if one zooms
out on a bipolar image, the result in image would reveal progressively less and
less information as the phases within the bipolar trace average out to
zero.
Therefore for most
viewing and interpretation purposes, one would likely choose either the
envelope-detected option or the halfwave-rectified
option. For most data archive and
transmission of the waveform data, one might choose the bipolar option.
One might produce
bit-reduced version of the envelope and halfwave, at
8 bits for example, and distribute these small but detailed versions to work
with and save the full bit-depth bipolar version for the archival purposes.
Compression parameters:
half wave rectified
Check off this box if you desire to create a half wave rectified version of input SEGY file. The resultant file will have an “_halfwave” suffix added to the root name of the input SEGY file.
This half wave rectified version will produce image very similar to the types of data recorded on EPC graphic recorders in the past. The sharpness of the phase in the waveforms will be retained. And again, like the envelope detected waveform, the half wave rectified image has a nonzero mean, and therefore will display well as one zooms in and out.
Compression
parameters: reversible transform
By default, the JPEG 2000 compression engine uses the 9 x 7 biorthogonal wavelet transforms, which are not precisely reversible. It is however, a relatively long filter and will provide better looking imagery than once generated with the shorter 5x3 reversible transform pairs.
The 5 x 3 transform pairs will produce a perfectly reversible encoding so that data can exactly reproduced when converted back into SEGY from JPEG 2000, at the cost of the visual quality.
Generally this option should only be used if there are real concerns about reconstructing an exact replica of the input SEGY file. Since the signal to noise ratio SEGY data is generally quite low, reconstruction errors are generally found below the noise threshold so that this concern is rarely one justified in real data sets.
Compression
parameters: make a local copy for processing
At the GSC, many of our source files sit on network attached storage boxes on the local network, and sometimes the I/O performance of these boxes leave something to be desired. If the speed of compression as concern, this option may be enabled so that a copy of the file will be made locally to speed up the process.
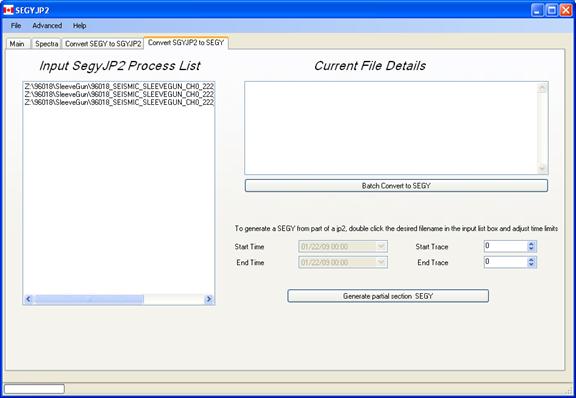
Convert JP2 to SEGY Tab Page:
Jp2-encoded SEGY files may be converted back into normal SEGY form. A list of files will be converted in btach mode back into SEGY form. If a subsection of a chosen file is desired, then double-click on the entry in the left-most list box and use the lower-right numeric entry box in order to choose new limits.
This conversion process cannot be applied to strictly image-type jp2 files that may been generated on a scanner for instance. These image files contain no encoded trace header information.
SEGY files that had been encoded with a band pass filter, an envelope detection or halfwave rectified will be reconstructed as floating point SEGY files regardless of their original format.
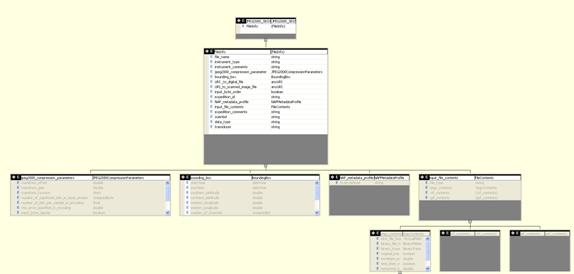
Appendix I - Storage and XML Schema
Trace data are stored in the image part of the JPEG2000 file in a compressed, variable bit length format.
SEGY file and trace header information and other related ancillary data (e.g., JPEG 2000 compression and bounding box data) are stored in a user-defined box within the JPEG 2000 file as gzipped XML with the box label, “sxml”.
Interpretation data are stored in a separate box as gzipped XML with the box label, “hxml”.
These schema are subject to change. However, effort will be made to make newer versions backwards compatible with the versions presented here.
The schema were created in Visual Studio 2005 and consequently, the schema may be used to generate class libraries for I/O using the utility, XSD.exe for C++, C# or Visual Basic. I believe Java and other languages offer similar functionality.
A class library (written in C++ for Visual Studio 2005) for manipulating boxes within JPEG 2000 files is given in Appendix 2 and is available from the author.
We have limited capacity to provide assistance and support to outside users.
Copies of these schema are located in the Program Files\NRCan\SEGYJP2 directory.
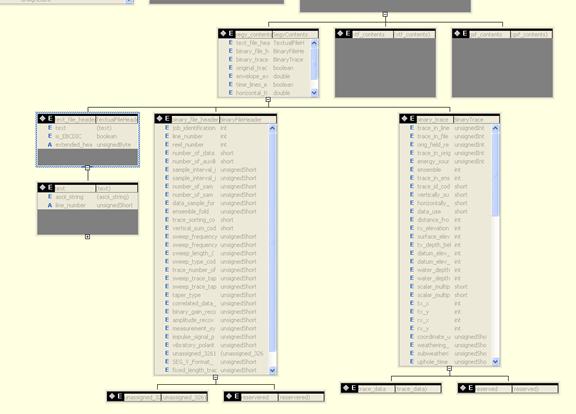
SEGY Header Schema
?xml version="1.0" encoding="utf-8"?>
<xs:schema xmlns:ns1="SEGY_rev_1" attributeFormDefault="unqualified" elementFormDefault="qualified" xmlns:xs="http://www.w3.org/2001/XMLSchema">
<xs:element name="JPEG2000_SEGY">
<xs:complexType>
<xs:sequence>
<xs:element name="FileInfo">
<xs:complexType>
<xs:all>
<xs:element name="file_name" type="xs:string" maxOccurs="1" minOccurs="1" />
<xs:element type="xs:string" name="instrument_type" maxOccurs="1" minOccurs="1" />
<xs:element name="instrument_comments" type="xs:string" minOccurs="0" maxOccurs="1" />
<xs:element name="jpeg2000_compression_parameters" type="JPEG2000CompressionParameters" maxOccurs="1" minOccurs="1">
</xs:element>
<xs:element name="bounding_box" type="BoundingBox" maxOccurs="1" minOccurs="1">
</xs:element>
<xs:element name="URI_x0020__to_digital_file" type="xs:anyURI" minOccurs="0" maxOccurs="1" />
<xs:element name="URI_to_scanned_image_file" type="xs:anyURI" minOccurs="0" maxOccurs="1" />
<xs:element name="input_byte_order" type="xs:boolean" minOccurs="1" maxOccurs="1" />
<xs:element name="expedition_id" type="xs:string" />
<xs:element name="NAP_metadata_profile" type="NAPMetadataProfile" minOccurs="0" maxOccurs="1" />
<xs:element name="input_file_contents" type="FileContents" minOccurs="1" maxOccurs="1" />
<xs:element name="expedition_comments" type="xs:string" maxOccurs="1" minOccurs="0" />
<xs:element name="scientist" type="xs:string" minOccurs="0" maxOccurs="1" />
<xs:element name="data_type" type="xs:string" />
<xs:element name="transducer" type="xs:string" />
</xs:all>
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:complexType name="BoundingBox">
<xs:sequence>
<xs:element name="start_x0020_time" type="xs:dateTime" minOccurs="1" maxOccurs="1" />
<xs:element name="end_x0020_time" type="xs:dateTime" maxOccurs="1" minOccurs="1" />
<xs:element name="southern_latititude" type="xs:double" maxOccurs="1" minOccurs="1" />
<xs:element name="northern_latititude" type="xs:double" maxOccurs="1" minOccurs="1" />
<xs:element name="western_longitude" type="xs:double" maxOccurs="1" minOccurs="1" />
<xs:element name="eastern_longitude" type="xs:double" maxOccurs="1" minOccurs="1" />
<xs:element name="number_of_channels" type="xs:unsignedInt" maxOccurs="1" minOccurs="1" />
<xs:element name="number_of_traces" type="xs:unsignedInt" maxOccurs="1" minOccurs="1" />
<xs:element name="minimum_trace_time" type="xs:int" maxOccurs="1" minOccurs="1" />
<xs:element name="maximum_trace_time_x0020_" type="xs:int" maxOccurs="1" minOccurs="1" />
<xs:element name="number_of_samples_per_trace" type="xs:unsignedInt" />
<xs:any />
</xs:sequence>
</xs:complexType>
<xs:complexType name="BinaryFileHeader">
<xs:sequence>
<xs:element name="job_identification_number" type="xs:int" />
<xs:element name="line_number" type="xs:int">
</xs:element>
<xs:element name="reel_number" type="xs:int" />
<xs:element name="number_of_data_traces_per_ensemble" type="xs:short" />
<xs:element name="number_of_auxiliary_traces_per_ensemble" type="xs:short" />
<xs:element name="sample_interval_in_microseconds" type="xs:unsignedShort" />
<xs:element type="xs:unsignedShort" name="sample_interval_in_usec_of_original_field_recording" />
<xs:element name="number_of_samples_per_data_trace" type="xs:unsignedShort" />
<xs:element name="number_of_samples_per_data_trace_for_original_field_recording" type="xs:unsignedShort" />
<xs:element name="data_sample_format_code" type="xs:unsignedShort" />
<xs:element name="ensemble_fold" type="xs:unsignedShort" />
<xs:element name="trace_sorting_code" type="xs:short" />
<xs:element name="vertical_sum_code" type="xs:short" />
<xs:element name="sweep_frequency_at_start__x0028_Hz_x0029_" type="xs:unsignedShort" />
<xs:element name="sweep_frequency_at_end__x0028_Hz_x0029_" type="xs:unsignedShort" />
<xs:element name="sweep_length__x0028_ms_x0029_" type="xs:unsignedShort" />
<xs:element name="sweep_type_code" type="xs:unsignedShort" />
<xs:element name="trace_number_of_sweep_channel" type="xs:unsignedShort" />
<xs:element name="sweep_trace_taper_length_in_milliseconds_at_start" type="xs:unsignedShort" />
<xs:element name="sweep_trace_taper_length_in_milliseconds_at_end" type="xs:unsignedShort" />
<xs:element name="taper_type" type="xs:unsignedShort" />
<xs:element name="correlated_data_traces" type="xs:unsignedShort" />
<xs:element name="binary_gain_recovered" type="xs:unsignedShort" />
<xs:element name="amplitude_recovery_method" type="xs:unsignedShort" />
<xs:element name="measurement_system" type="xs:unsignedShort" />
<xs:element name="impulse_signal_polarity" type="xs:unsignedShort" />
<xs:element name="vibratory_polarity_code" type="xs:unsignedShort" />
<xs:element name="unassigned_3261_3500" minOccurs="0" maxOccurs="1">
<xs:simpleType>
<xs:restriction base="xs:base64Binary">
<xs:length value="240" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name="SEG_Y_Format_Revision_Number" type="xs:unsignedShort" default="0" minOccurs="0" maxOccurs="1" />
<xs:element name="fixed_length_trace_flag" type="xs:unsignedShort" default="0" minOccurs="0" maxOccurs="1" />
<xs:element name="number_of_extended_Textual_files" type="xs:short" default="0" minOccurs="0" maxOccurs="1" />
<xs:element name="reservered">
<xs:simpleType>
<xs:restriction base="xs:base64Binary">
<xs:length value="94" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
</xs:complexType>
<xs:complexType name="BinaryTrace">
<xs:sequence>
<xs:element name="trace_in_line" type="xs:unsignedInt" minOccurs="0" maxOccurs="1" />
<xs:element name="trace_in_file" type="xs:unsignedInt" minOccurs="0" maxOccurs="1" />
<xs:element name="orig_field_record" type="xs:unsignedInt" minOccurs="0" maxOccurs="1" />
<xs:element name="trace_in_orig_field_record" type="xs:unsignedInt" minOccurs="0" maxOccurs="1" />
<xs:element name="energy_source_point" type="xs:unsignedInt" minOccurs="0" maxOccurs="1" />
<xs:element name="ensemble" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="trace_in_ensemble" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="trace_id_code" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="vertically_summed_traces" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="horizontally_summed_traces" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="data_use" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="distance_from_tx_to_rx" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="rx_elevation_" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="surface_elev_at_tx" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="tx_depth_below_surface" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="datum_elev_at_rx" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="datum_elev_at_tx" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="water_depth_at_tx" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="water_depth_at_rx" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="scalar_multiplier_69-70" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="scalar_multiplier_71-72" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="tx_x" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="tx_y" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="rx_x" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="rx_y" type="xs:int" minOccurs="0" maxOccurs="1" />
<xs:element name="coordinate_units" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="weathering_velocity" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="subweathering_velocity" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="uphole_time_at_tx" type="xs:unsignedShort" minOccurs="0" maxOccurs="1 " />
<xs:element name="uphole_time_at_rx" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="tx_static_correction_" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="rx_static_correction_" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="total_static_correction_" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="lag_time_A" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="lag_time_B" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="delay_record_time" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="mute_start_time" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="mute_end_time" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="number_of_samples_in_trace" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sample_interval__x0028_usec_x0029_" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="gain_type_of_field_instruments" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="instrument_gain_constant" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="instrument_initial_gain" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="correlated" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sweep_freq_start" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sweep_freq_end" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sweep_length__x0028_msec_x0029_" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sweep_type" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sweep_taper_length_start" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sweep_taper_length_end" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="taper_type" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="antialias_filter_frequency" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="antialias_filter_slope" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="notch_filter_freq" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="notch_filter_slope" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="low-cut_freq" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="high_cut_freq" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="low-cut_slope" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="high-cut_slope" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="year_data_recorded" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="day" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="hr" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="min" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="sec" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="msec" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="time_weighting_factor" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="group_number_of_roll_sw_1" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="group_number_of_tr1" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="group_number_of_last_trace" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="gap_size" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="over_travel" type="xs:unsignedShort" minOccurs="0" maxOccurs="1" />
<xs:element name="trace_data" minOccurs="0" maxOccurs="1">
<xs:simpleType>
<xs:restriction base="xs:base64Binary">
<xs:maxLength value="64000" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:element name="taper" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="TriggerInterval" type="xs:short" minOccurs="0" maxOccurs="1" />
<xs:element name="reserved">
<xs:simpleType>
<xs:restriction base="xs:base64Binary">
<xs:length value="58" />
</xs:restriction>
</xs:simpleType>
</xs:element>
<xs:any />
</xs:sequence>
<xs:attribute name="is_reference_header" type="xs:boolean" default="false" />
<xs:attribute name="trc_num" type="xs:int" />
</xs:complexType>
<xs:complexType name="TextualFileHeader">
<xs:sequence>
<xs:element name="text" minOccurs="40" maxOccurs="40">
<xs:complexType>
<xs:sequence>
<xs:element name="ascii_string">
<xs:simpleType>
<xs:restriction base="xs:string">
<xs:length value="80" />
</xs:restriction>
</xs:simpleType>
</xs:element>
</xs:sequence>
<xs:attribute name="line_number" type="xs:unsignedShort" />
</xs:complexType>
</xs:element>
<xs:element name="is_EBCDIC" type="xs:boolean" />
</xs:sequence>
<xs:attribute name="extended_header_number" type="xs:unsignedByte" />
</xs:complexType>
<xs:complexType name="JPEG2000CompressionParameters">
<xs:sequence>
<xs:element name="transform_offset" type="xs:double" />
<xs:element name="transform_gain" type="xs:double" />
<xs:element name="transform_function" type="xs:short" />
<xs:element name="number_of_significant_bits_in_input_stream" type="xs:unsignedByte" />
<xs:element type="xs:float" name="number_of_bits_per_sample_in_encoding" />
<xs:element name="rms_error_specified_in_encoding" type="xs:double" />
<xs:element name="input_trace_bipolar" type="xs:boolean" />
<xs:element name="significant_bits_at_top_of_word" type="xs:boolean" />
<xs:element name="minimum_trace_delay_x0020__x0028_msec_x0029_" type="xs:int" />
<xs:element name="padded_trace_length" type="xs:unsignedInt">
</xs:element>
<xs:element name="jp2000_wavelet_transform" type="xs:unsignedByte" />
</xs:sequence>
</xs:complexType>
<xs:complexType name="SEGYContents">
<xs:sequence>
<xs:element name="text_file_header" type="TextualFileHeader" minOccurs="1" />
<xs:element name="binary_file_header" type="BinaryFileHeader" minOccurs="1" maxOccurs="1" />
<xs:element name="binary_trace" type="BinaryTrace" minOccurs="1" maxOccurs="unbounded" />
<xs:element name="original_trace_converted_to_envelope" type="xs:boolean" minOccurs="0" maxOccurs="1" default="false" />
<xs:element name="envelope_exponent" type="xs:double" minOccurs="0" maxOccurs="1" default="1.0" />
<xs:element name="time_lines_embedded_in_trace_data" type="xs:boolean" default="false" maxOccurs="1" minOccurs="0" />
<xs:element name="horizontal_time_line_separation_msec" type="xs:double" minOccurs="0" maxOccurs="1" default="250" />
<xs:element name="vertical_time_line_separation_sec" type="xs:double" minOccurs="0" maxOccurs="1" default="600." />
<xs:element name="original_trace_converted_to_half_wave" type="xs:boolean" default="false" minOccurs="0" maxOccurs="1" />
</xs:sequence>
</xs:complexType>
<xs:complexType name="NAPMetadataProfile">
<xs:sequence>
<xs:element name="to_x0020_be_x0020_defined" type="xs:string" />
</xs:sequence>
</xs:complexType>
<xs:complexType name="FileContents">
<xs:sequence>
<xs:element name="file_type" type="xs:string" />
<xs:element name="SEGY_contents" type="SEGYContents" minOccurs="0" maxOccurs="1" />
<xs:element name="xtf_contents" minOccurs="0" maxOccurs="1">
<xs:complexType>
<xs:sequence />
</xs:complexType>
</xs:element>
<xs:element name="gsf_contents" minOccurs="0" maxOccurs="1">
<xs:complexType>
<xs:sequence />
</xs:complexType>
</xs:element>
</xs:sequence>
</xs:complexType>
</xs:schema>
Horizon Schema
Interpretations are
stored in a separate box in structured XML. The intent here is to separate
interpretations from the data, but keep them packaged together.
Although it has not
yet been implemented, it is intended to offer the capacity to have multiple
interpretations included with a given data set, offering the capacity to
exchange and reinterpret data sets in the lab and over the Internet.
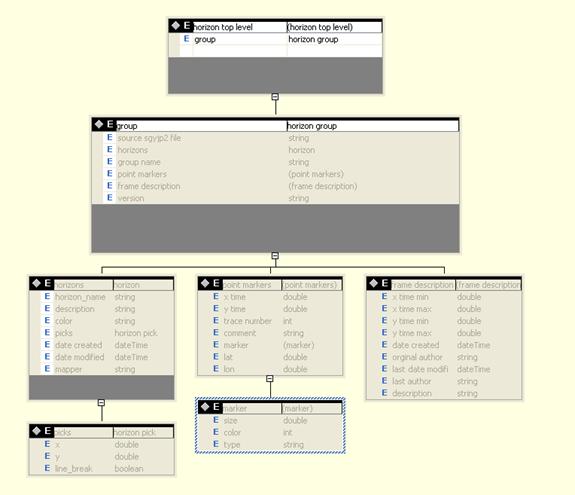
In the following
schema, all X coordinates are sequential trace numbers and all Y coordinates
are two-way travel times in milliseconds.
<?xml version="1.0" encoding="utf-8"?>
<xs:schema id="horizons" targetNamespace="http://tempuri.org/horizons.xsd" elementFormDefault="qualified" xmlns="http://tempuri.org/horizons.xsd" xmlns:mstns="http://tempuri.org/horizons.xsd" xmlns:xs="http://www.w3.org/2001/XMLSchema">
<xs:complexType name="horizon_x0020_pick">
<xs:sequence>
<xs:element name="x" type="xs:double" />
<xs:element name="y" type="xs:double" />
<xs:element name="line_break" type="xs:boolean" default="false" minOccurs="0" maxOccurs="1" />
</xs:sequence>
</xs:complexType>
<xs:complexType name="horizon">
<xs:sequence>
<xs:element name="horizon_name" type="xs:string" minOccurs="1" maxOccurs="1" />
<xs:element name="description" type="xs:string" minOccurs="0" />
<xs:element name="color" type="xs:string" minOccurs="1" maxOccurs="1" />
<xs:element name="picks" type="horizon_x0020_pick" minOccurs="0" maxOccurs="unbounded" />
<xs:element name="date_x0020_created" type="xs:dateTime" minOccurs="0" />
<xs:element name="date_x0020_modified" type="xs:dateTime" />
<xs:element name="mapper" type="xs:string" minOccurs="0" />
</xs:sequence>
</xs:complexType>
<xs:complexType name="horizon_x0020_group">
<xs:sequence>
<xs:element name="source_x0020_sgyjp2_x0020_file" type="xs:string" />
<xs:element name="horizons" type="horizon" minOccurs="1" maxOccurs="unbounded" />
<xs:element name="group_x0020_name" type="xs:string" />
<xs:element name="point_x0020_markers" minOccurs="0" maxOccurs="unbounded">
<xs:complexType>
<xs:sequence>
<xs:element name="x_x0020_time" type="xs:double" />
<xs:element name="y_x0020_time" type="xs:double" />
<xs:element name="trace_x0020_number" type="xs:int" />
<xs:element name="comment" type="xs:string" />
<xs:element name="marker" minOccurs="0" maxOccurs="1">
<xs:complexType>
<xs:sequence>
<xs:element name="size" type="xs:double" />
<xs:element name="color" type="xs:int" />
<xs:element name="type" type="xs:string" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name="lat" type="xs:double" minOccurs="0" maxOccurs="1" />
<xs:element name="lon" type="xs:double" minOccurs="0" maxOccurs="1" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name="frame_x0020_description" maxOccurs="unbounded" minOccurs="0">
<xs:complexType>
<xs:sequence>
<xs:element name="x_x0020_time_x0020_min" type="xs:double" />
<xs:element name="x_x0020_time_x0020_max" type="xs:double" />
<xs:element name="y_x0020_time_x0020_min" type="xs:double" />
<xs:element name="y_x0020_time_x0020_max" type="xs:double" />
<xs:element name="date_x0020_created" type="xs:dateTime" />
<xs:element name="orginal_x0020_author" type="xs:string" />
<xs:element name="last_x0020_date_x0020_modified" type="xs:dateTime" />
<xs:element name="last_x0020_author" type="xs:string" />
<xs:element name="description" type="xs:string" />
</xs:sequence>
</xs:complexType>
</xs:element>
<xs:element name="version" type="xs:string" minOccurs="0" maxOccurs="1" />
</xs:sequence>
</xs:complexType>
<xs:element name="horizon_x0020_top_x0020_level">
<xs:complexType>
<xs:sequence>
<xs:element name="group" type="horizon_x0020_group" minOccurs="1" maxOccurs="unbounded" />
</xs:sequence>
</xs:complexType>
</xs:element>
</xs:schema>
Routines to Manipulate JPEG2000
Box Data
JPEG2000_Manage.h ( Visual
Studio 2005 .NET)
#pragma once
using namespace
System;
using namespace
System::IO;
using namespace
System::IO::Compression;
using namespace
System::Runtime::InteropServices;
using namespace
System::Xml;
using namespace
System::Xml::Serialization;
using namespace
System::Collections;
ref class jp2_box_data
{
public:
unsigned int position;
unsigned int box_length;
String^ box_label;
Boolean xl_box;
Boolean zero_length_box;
};
#define IO_BUFFER_LENGTH
5000000; // define a 5 mbyte io buffer
ref class
JPEG2000_Manage
{
public:
JPEG2000_Manage(void);
property String^ filename;
property unsigned int box_length;
property Boolean xl_box;
property Boolean zero_length_box;
property FileStream^
filestream;
property BinaryReader^
binaryreader;
property BinaryWriter^
binarywriter;
Boolean has_sxml_content(void); // check to see if
it has SEGY formatted xml content
// open up a file for read write
Boolean open(void); // file name preset with property filename;
Boolean open(String^);
Boolean open(Stream^ s);
Boolean close(void);
Boolean is_valid_jp2(void); // looks for
"jP " header box
// get the next box header - returns box label as String^ ;
box length is in this->box_length; returns null string if failed or EOF
// leave the cuurent stream of file at start of box
contents
String^ get_next_box_header(void);
String^ get_next_box_header(Stream^ s);
// skip over the box contents leaving the file postion at
start of next jp2000 box
Boolean skip_box_contents(void);
Boolean skip_box_contents(Stream^ s);
// set filestream
void go_to_top_of_file(void);
// checks to see if at least one box with a given box_label
exists
bool find_box(String^ box_label);
// fix zero length last box in file
bool fix_zero_length_box();
// write an serializable object to an xml file
static Boolean write_object_to_xml_text_file(Object^ input_object,
String^ xml_text_file_name);
static Object^ read_object_from_xml_text_file(System::Type^ input_object_type, String^
xml_text_file_name);
// write a serializable object to a gzipped xml file
static Boolean write_object_to_xml_gzip_file(Object^ input_object,
String^ xml_gzip_file_name);
static Object^ read_object_from_xml_gzip_file(System::Type^ output_object_type, String^
xml_gzip_file_name);
static System::Void read_object_from_xml_gzip_file(Object^ output_object,
String^ xml_gzip_file_name);
// write an xml version of an object in gzipped format to a
stream
Boolean write_object_to_xml_gzip_stream(Object^ input_object,
Stream^ ms);
// write an xml version of an object in text format to a
stream
Boolean write_object_to_xml_stream(Object^ input_object,
Stream^ ms);
// write serializable object in gzipped xml
format to a jp2000 compliant box at the current stream position
Boolean write_compressed_xml_box(Object^ input_object,
String^ box_label,
String^ gzip_xml_box_file_name);
// write serializable object in gzipped xml format to a
jp2000 compliant box at the current stream position
Boolean write_compressed_xml_box(Object^ input_object,
String^ box_label,
Stream^ s);
// write serializable object in readable xml format to a jp2000 compliant
box in a file
Boolean write_xml_box(Object^ input_object,
String^ box_label,
String^ xml_box_file_name);
// write serializable object in readable xml format to a jp2000 compliant
box at the current stream position
Boolean write_xml_box(Object^ input_object,
String^ box_label,
Stream^ s);
// Read an xml object from a stream whether compressed or
not
Object^ read_xml_box(System::Type^ input_object_type, String^
box_label , Stream^
s);
// Copy a jp2000 compliant
box from one stream to another
Boolean copy_box(Stream^ input_stream,
Stream^ output_stream);
// get a summary list of jp2000 boxes in current file -
creates an arraylist of type jp2_box_data
ArrayList^ get_box_data(void);
// compress current jp2 file
Boolean compress_existing_sxml_box(String^ new_file_name);
// does the file contain compressed xml content
Boolean has_compressed_sxml_content(void);
private:
Boolean input_file_is_open;
array<unsigned char,1>^ box_type;
array<unsigned char,1>^ box_XL;
void Write_Box_Header_to_MemoryStream(String ^box_label, BinaryWriter ^bw, MemoryStream ^ms);
};
JPEG2000_Manage.cpp (
Visual Studio 2005 .NET)
#include "StdAfx.h"
#include "stdio.h"
#include "JPEG2000_Manage.h"
#include "SEGY.h"
// sxml header class
// utilities
System::UInt32 swap(UInt32 x)
{
// implement a swap
array<unsigned char,1>^ c = BitConverter::GetBytes(x);
unsigned char tmp;
tmp = c[0];
c[0] = c[3];
c[3] = tmp;
tmp = c[1];
c[1] = c[2];
c[2] = tmp;
return BitConverter::ToUInt32(c,0);
}
System::UInt16 swap(UInt16 x)
{
// implement a swap
array<unsigned char,1>^ c = BitConverter::GetBytes(x);
unsigned char tmp;
tmp = c[0];
c[0] = c[1];
c[1] = tmp;
return BitConverter::ToUInt16(c,0);
}
System::Int16 swap(Int16 x)
{
// implement a swap
array<unsigned char,1>^ c = BitConverter::GetBytes(x);
unsigned char tmp;
tmp = c[0];
c[0] = c[1];
c[1] = tmp;
return BitConverter::ToInt16(c,0);
}
JPEG2000_Manage::JPEG2000_Manage(void)
{
this->filestream
= nullptr;
this->binaryreader
= nullptr;
this->filename = nullptr;
this->input_file_is_open
= false;
this->box_length
= -1;
}
System::Boolean
JPEG2000_Manage::has_sxml_content(void)
{
return this->find_box("sxml");
}
System::Boolean
JPEG2000_Manage::open(void)
{
if( this->filename == nullptr)
return false;
this->input_file_is_open=
false;
try
{
this->filestream
= File::Open(this->filename,FileMode::OpenOrCreate);
}
catch (System::Exception^ )
{
return false;
}
try
{
this->binaryreader
= gcnew BinaryReader(this->filestream);
}
catch ( System::Exception^)
{
return false;
}
try
{
this->binarywriter=
gcnew BinaryWriter(this->filestream);
}
catch ( System::Exception^)
{
return false;
}
this->input_file_is_open
= true;
return true;
}
System::Boolean
JPEG2000_Manage::open(String^ input_name)
{
this->filename = String::Copy(input_name); // set
filename
return this->open();
}
System::Boolean
JPEG2000_Manage::open(Stream^ s)
{
this->filestream
= (FileStream^) s;
try
{
this->binaryreader
= gcnew BinaryReader(this->filestream);
}
catch ( System::Exception^)
{
return false;
}
try
{
this->binarywriter=
gcnew BinaryWriter(this->filestream);
}
catch ( System::Exception^)
{
return false;
}
this->filename = "Stream input"; //
set filename
return true;
}
String^ JPEG2000_Manage::get_next_box_header(void)
{
return this->get_next_box_header(this->filestream);
}
String^ JPEG2000_Manage::get_next_box_header(Stream
^s)
{
if ( s->Position >= s->Length ) return nullptr; // at eof
BinaryReader^
binaryreader = gcnew BinaryReader(s);
// read the box header bytes
// jp2 are
big-endian
box_length = swap(binaryreader->ReadUInt32()) ;
// set zero box
length flag
this->zero_length_box = false;
if( !box_length)
{
this->zero_length_box = true;
this->box_length = s->Length - s->Position + 4;
}
//
read box type
box_type = binaryreader->ReadBytes(4);
this->xl_box = false;
// read next eight
bytes - note first 4 bytes should be all zeroes for 32 bit file sizes
if ( box_length == 1 )
{
this->xl_box = true;
box_XL = binaryreader->ReadBytes(8);
// byte swap part of
word
for ( int i = 0; i < 4 ; i++ ) {
unsigned char tmp = box_XL[i];
if ( tmp != NULL )
{
// high bytes should
be zero for 32 bit files lengths
return nullptr;
}
box_XL[i] = box_XL[7-i];
box_XL[7-i] = tmp;
}
// limit to 32 bit
lengths; adjust length for preamble bytes
box_length =Convert::ToUInt32( BitConverter::ToUInt64(box_XL,0)) ;
}
// copy box_type
into holding array - this is a real hack!!
unsigned char tmp[5];
for ( int i = 0; i < 4; i++) tmp[i] = box_type[i];
tmp[4] = 0;
String^ name = Marshal::PtrToStringAnsi( (IntPtr) tmp);
return name;
}
Boolean JPEG2000_Manage::close(void)
{
this->filestream->Close();
this->input_file_is_open
= false;
return true;
}
Boolean JPEG2000_Manage::is_valid_jp2(void)
{
this->go_to_top_of_file();
// make sure the first box type is jp2/jp2
if(String::Compare(this->get_next_box_header(),"jP ") == 0 )
{
this->go_to_top_of_file();
return true;
}
return false;
}
void JPEG2000_Manage::go_to_top_of_file(void)
{
if( this->input_file_is_open) this->filestream->Seek(0,SeekOrigin::Begin);
}
Boolean JPEG2000_Manage::skip_box_contents(void)
{
return this->skip_box_contents(this->filestream);
}
Boolean JPEG2000_Manage::skip_box_contents(Stream^
s)
{
// advance to end of box contents
unsigned int new_position;
if( this->box_length > 0 )
{
new_position = s->Position + this->box_length
- 8;
if ( this->xl_box) new_position
-= 8; // adjust skip for extra 8 bytes in xl box
if ( new_position
>= s->Length)
{
// EOF
s->Position = s->Length ;
return false;
}
else {
s->Position = new_position ;
}
return true;
}
else
{
// box extends to end of file - zero length trace
// find current file position
int box_start = s->Position - 8;
s->Position = s->Length; // move to EOF
this->box_length
= s->Position
- box_start;
return true;
}
}
bool JPEG2000_Manage::fix_zero_length_box()
{
ArrayList^ boxdata
= this->get_box_data();
// last box
this->filestream->Position = ((jp2_box_data^)boxdata[boxdata->Count - 1])->position;
if ( ((jp2_box_data^)boxdata[boxdata->Count - 1])->zero_length_box
)
{
unsigned int swapped = swap(((jp2_box_data^)boxdata[boxdata->Count -
1])->box_length);
BinaryWriter^ bw = gcnew BinaryWriter(this->filestream);
bw->Write( swapped);
}
// done
return true;
}
bool JPEG2000_Manage::find_box(String^ boxname)
{
if( !this->input_file_is_open ) return
false;
if( !this->is_valid_jp2()) return
false;
// loop through boxes and box contents
while ( String^ name = this->get_next_box_header())
{
if ( String::Compare(name,boxname) == 0 )
{
this->go_to_top_of_file();
return true;
}
this->skip_box_contents();
if( this->filestream->Position
== this->filestream->Length - 1 ) // eof
condition
{
this->go_to_top_of_file();
return false;
}
}
return false;
}
// this routine writes
an object to a XML text file
Boolean JPEG2000_Manage::write_object_to_xml_text_file(Object^ input_object,
String^ xml_text_file_name)
{
XmlSerializer^ ser
= gcnew XmlSerializer(input_object->GetType());
XmlTextWriter^ writer
= gcnew XmlTextWriter(
xml_text_file_name, System::Text::Encoding::UTF8);
ser->Serialize(writer,input_object);
writer->Close();
return true;
}
Object^ JPEG2000_Manage::read_object_from_xml_text_file(System::Type^ input_object_type, String^
xml_text_file_name)
{
Object^ output_object
= nullptr;
XmlSerializer^ ser
= gcnew XmlSerializer(input_object_type);
XmlTextReader^ reader
= gcnew XmlTextReader(xml_text_file_name);
output_object = ser->Deserialize(reader);
reader->Close();
return output_object;
}
Object^
JPEG2000_Manage::read_object_from_xml_gzip_file(System::Type^ output_object_type, String^
xml_gzip_file_name)
{
// we should be a the start of the desired box
// check the next two bytes if they are 1f8b then its a
gzip compressed box
Object^ output_object = nullptr;
FileStream^ s = gcnew FileStream(xml_gzip_file_name,System::IO::FileMode::Open);
XmlSerializer^ ser
= gcnew XmlSerializer(output_object_type);
GZipStream^ gs = gcnew GZipStream(s,CompressionMode::Decompress);
output_object
= ser->Deserialize(gs);
gs->Close();
s->Close();
return output_object;
}
System::Void JPEG2000_Manage::read_object_from_xml_gzip_file(Object^ output_object,
String^ xml_gzip_file_name)
{
// we should be a the start of the desired box
// check the next two bytes if they are 1f8b then its a
gzip compressed box
FileStream^ s = gcnew FileStream(xml_gzip_file_name,System::IO::FileMode::Open);
XmlSerializer^ ser
= gcnew XmlSerializer(output_object->GetType());
GZipStream^ gs = gcnew GZipStream(s,CompressionMode::Decompress);
output_object
= ser->Deserialize(gs);
return;
}
Boolean JPEG2000_Manage::write_object_to_xml_gzip_file(Object^ input_object,
String^ xml_gzip_file_name)
{
FileStream^ fs
= nullptr;
try
{
fs = gcnew FileStream(xml_gzip_file_name,FileMode::Create);
}
catch ( System::Exception^ e)
{
return false;
}
GZipStream ^ compressedzipStream
= gcnew GZipStream(fs,CompressionMode::Compress,true );
XmlSerializer^ ser
= gcnew XmlSerializer(input_object->GetType());
XmlTextWriter^ writer
= gcnew XmlTextWriter(
compressedzipStream, System::Text::Encoding::UTF8);
ser->Serialize(writer,input_object);
writer->Close();
compressedzipStream->Close();
fs->Close();
return true;
}
Boolean JPEG2000_Manage::write_object_to_xml_gzip_stream(Object^ input_object,
Stream^ ms)
{
GZipStream ^ compressedzipStream
= gcnew GZipStream(ms,CompressionMode::Compress,true );
this->write_object_to_xml_stream(input_object,compressedzipStream);
compressedzipStream->Close();
return true;
}
Boolean JPEG2000_Manage::write_object_to_xml_stream(Object^
input_object, Stream^
ms)
{
XmlSerializer^ ser
= gcnew XmlSerializer(input_object->GetType());
XmlTextWriter^ writer
= gcnew XmlTextWriter(
ms, System::Text::Encoding::UTF8);
ser->Serialize(writer,input_object);
return true;
}
Boolean JPEG2000_Manage::write_compressed_xml_box(Object^
input_object, String^
box_label, String^
gzip_xml_box_file_name)
{
FileStream^ ms = gcnew FileStream(gzip_xml_box_file_name,FileMode::Create);
BinaryWriter^
bw = gcnew BinaryWriter(ms);
// write 8 byte
preamble to box
bw->Write((int)0);
char* str2 = (char*)(void*)Marshal::StringToHGlobalAnsi(box_label);
for( int i = 0; i < 4; i++) bw->Write((unsigned char) str2[i]);
// write gzipped
contents
this->write_object_to_xml_gzip_stream(input_object,ms);
// update length of
box
unsigned int ms_size = ms->Length;
ms->Position = 0;
bw->Write(swap(ms_size));
// close up
bw->Close();
return true;
}
Boolean JPEG2000_Manage::write_xml_box(Object^
input_object, String^
box_label, String^
xml_box_file_name)
{
FileStream^ ms = gcnew FileStream(xml_box_file_name,FileMode::Create);
BinaryWriter^
bw = gcnew BinaryWriter(ms);
// write 8 byte
preamble to box
bw->Write((int)0);
char* str2 = (char*)(void*)Marshal::StringToHGlobalAnsi(box_label);
for( int i = 0; i < 4; i++) bw->Write((unsigned char) str2[i]);
// write contents
.......
this->write_object_to_xml_stream(input_object,ms);
// update length of
box
unsigned int ms_size = ms->Length;
ms->Position = 0;
bw->Write(swap(ms_size));
// close up
bw->Close();
return true;
}
void JPEG2000_Manage::Write_Box_Header_to_MemoryStream(String ^box_label, BinaryWriter ^bw, MemoryStream ^ms)
{
// write preamble to stream - note that a memory stream
< 2 Gbyte
bw->Write(swap((unsigned int)(ms->Length + 8)));
char* str2 = (char*)(void*)Marshal::StringToHGlobalAnsi(box_label);
for( int i = 0; i < 4; i++) bw->Write((unsigned char) str2[i]);
}
Boolean JPEG2000_Manage::write_xml_box(Object^
input_object, String^
box_label, Stream^
s)
{
BinaryWriter^
bw = gcnew BinaryWriter(s);
MemoryStream^
ms = gcnew MemoryStream();
// put text xml in a
memory stream
this->write_object_to_xml_stream(input_object,ms);
Write_Box_Header_to_MemoryStream(box_label, bw, ms);
// dump memory stream to parent stream
ms->WriteTo(s);
return true;
}
Boolean JPEG2000_Manage::write_compressed_xml_box(Object^
input_object, String^
box_label, Stream^
s)
{
BinaryWriter^
bw = gcnew BinaryWriter(s);
MemoryStream^
ms = gcnew MemoryStream();
// put gzip xml in a
memory stream
this->write_object_to_xml_gzip_stream(input_object,ms);
Write_Box_Header_to_MemoryStream(box_label, bw, ms);
// dump memory stream to parent stream
ms->WriteTo(s);
return true;
}
ArrayList^ JPEG2000_Manage::get_box_data(void)
{
ArrayList^ r = gcnew ArrayList();
this->go_to_top_of_file();
while(this->filestream->Position
< this->filestream->Length)
{
jp2_box_data^ bd = gcnew jp2_box_data();
bd->position = this->filestream->Position;
bd->box_label = this->get_next_box_header();
bd->box_length =
this->box_length;
bd->xl_box = this->xl_box;
bd->zero_length_box
= this->zero_length_box;
r->Add(bd);
this->skip_box_contents();
}
return r;
}
// Read an xml object
from a stream whether compressed or not
Object^ JPEG2000_Manage::read_xml_box(System::Type^ input_object_type,
String^ box_label
, Stream^ s)
{
Object^ output_object
= nullptr;
while ( String::Compare(this->get_next_box_header(s),
box_label) != 0 )
{
this->skip_box_contents(s);
if( s->Position >=
s->Length )return nullptr;
}
// we should be a the start of the desired box
// check the next two bytes if they are 1f8b then its a
gzip compressed box
BinaryReader^ br = gcnew BinaryReader(s);
unsigned char c1 = br->ReadByte();
unsigned char c2 = br->ReadByte();
s->Position -=
2; // back up
// read box contents
MemoryStream^ ms ;
if( this->xl_box )
{
ms = gcnew MemoryStream(br->ReadBytes(this->box_length-16));
}
else
{
ms = gcnew MemoryStream(br->ReadBytes(this->box_length-8));
}
XmlSerializer^ ser
= gcnew XmlSerializer(input_object_type);
if ( c1 == 31
&& c2 == 139 )
{
// this is gzipped xml
GZipStream^ gs = gcnew GZipStream(ms,CompressionMode::Decompress);
output_object
= ser->Deserialize(gs);
}
else {
output_object
= ser->Deserialize(ms);
}
return output_object;
}
Boolean JPEG2000_Manage::copy_box(Stream^ is, Stream^ os)
{
// straight copy of one box from one stream to another
// read box header
String^ is_box_label
= this->get_next_box_header(is);
if( this->xl_box)
{
is->Position -=
16; // xl box
}
else {
is->Position -=
8; // back up
}
if ( is_box_label ==
nullptr ) return
false;
// total length of box
unsigned int is_length = this->box_length;
if( this->box_length == 0 )
{
// this is the last box in the file
is_length = is->Length - is->Position;
}
// assign an io buffer;
unsigned int buffer_length = IO_BUFFER_LENGTH;
if( buffer_length
> is_length
) buffer_length = is_length ;
array<unsigned char>^ buffer = gcnew array<unsigned char>(buffer_length);
// number of reads
unsigned int nread_full = is_length/buffer_length;
for ( int i = 0; i < nread_full; i++ )
{
is->Read(buffer,0,buffer_length);
os->Write(buffer,0,buffer_length);
}
unsigned int nread_partial = is_length
% buffer_length;
if( nread_partial )
{
is->Read(buffer,0,nread_partial);
os->Write(buffer,0,nread_partial);
}
return true;
}
Boolean JPEG2000_Manage::has_compressed_sxml_content(void)
{
unsigned int save_position = this->filestream->Position;
ArrayList^ bd = this->get_box_data();
// get input box info
// look for sxml header
for ( int i = 0; i < bd->Count; i++ )
{
jp2_box_data^ b = (jp2_box_data^)bd[i];
if ( String::Compare(b->box_label,"sxml")
== 0 )
{
// found sxml tag
this->filestream->Position = b->position + 8;
if( b->xl_box ) this->filestream->Position
+= 8; // xl box
// next two characters are gzip magic numbers
unsigned char c1 = this->binaryreader->ReadByte();
unsigned char c2 = this->binaryreader->ReadByte();
if ( c1 == 31
&& c2 == 139 )
{
this->filestream->Position = save_position;
return true;
}
}
}
this->filestream->Position = save_position;
return false;
}
Boolean
JPEG2000_Manage::compress_existing_sxml_box(String^
new_file_name)
{
try
{
JPEG2000_Manage^ newjp2
= gcnew JPEG2000_Manage();
newjp2->open(new_file_name); // open a
blank file
ArrayList^ bd = this->get_box_data();
// get input box info
this->go_to_top_of_file();
// rewind existing file
for ( int i = 0; i < bd->Count; i ++ ) // cycle through
boxes
{
jp2_box_data^
bdi = (::jp2_box_data^)
bd[i];
if ( String::Compare(bdi->box_label,"sxml") == 0 )
{
JPEG2000_SEGY^
jp2sgy = gcnew
JPEG2000_SEGY() ;
jp2sgy = (JPEG2000_SEGY^) this->read_xml_box(jp2sgy->GetType(),"sxml",this->filestream);
//rewrite in a compressed
format
this->write_compressed_xml_box(jp2sgy,"sxml",newjp2->filestream);
} else {
this->copy_box(this->filestream, newjp2->filestream);
}
}
// insure output file has not zero length boxes
newjp2->fix_zero_length_box();
newjp2->close();
}
catch ( System::Exception^
)
{
return false;
}
return true;
}
Appendix 2: What is so special
about JPEG 2000
JPEG 2000 should
definitely not be confused with the commonly-used file format JPEG. It
represents a total reengineering of the way images and data can be packaged,
viewed, annotated,
and disseminated over Internet channels.
Although it is outside
the bounds of this document to fully discuss this framework, some of the
important and salient points of this approach to image storage of compression
are listed below:
- Open file standard ISO/IEC
15444-1:2000
- Wavelet
based, multiresolution representation
- Up to 38
bit signed data – not just images
- Up to
16,000 planes/channels
- Entropy-based
(MQ) bit-plane encoding (save 20 bits instead of 32, white space costs
almost nothing)
- Lossless/lossy encoding - harmonic distortion for lossy compression
- Flexible
file format –XML-aware, UUID
defined boxes
- Random access to ROI, transcoding, quality layers,etc
- Internet ready : JPIP => low
bandwidth optimized
- Industry support: e.g., Lizardtech, Adobe Photoshop
- Custom wavelet transforms